Differentially Active Gene Set Analysis
Seongyong_Park
2020-11-27
dagsanalysis.RmdDifferentially Active Gene Set Analysis with conventional methods
We often find differentially active gene sets in microarray dataset using some of well known methods. We can also apply overlap statistics for this purpose. Here I compared ovltools with GSVA, ssGSEA, and PLAGE.
suppressPackageStartupMessages(library(golubEsets))
suppressPackageStartupMessages(library(GSVA))
## Load Dataset
suppressPackageStartupMessages(library(dplyr))
data("Golub_Train")
data("Golub_Test")
suppressPackageStartupMessages(library(hu6800.db))
## inspaect
gex_train = Golub_Train@assayData$exprs
gex_test = Golub_Test@assayData$exprs
label_train = Golub_Train@phenoData@data$ALL.AML
label_test = Golub_Test@phenoData@data$ALL.AML
gene_tbl = data.frame(probe_id = names(as.list(hu6800ENTREZID)), Entrez=unlist(as.list(hu6800ENTREZID), use.names = F))
gene_tbl = gene_tbl[complete.cases(gene_tbl),]
## Mapping Gene IDs
gex_train = gex_train %>% as.data.frame() %>% mutate(probe_id = rownames(gex_train)) %>% inner_join(gene_tbl, by="probe_id") %>%
dplyr::select(-probe_id) %>% group_by(Entrez) %>% summarise_all(list(mean)) %>% as.data.frame()
rownames(gex_train) = gex_train$Entrez
gex_train = gex_train %>% dplyr::select(-Entrez)
## Mapping Gene IDs
gex_test = gex_test %>% as.data.frame() %>% mutate(probe_id = rownames(gex_test)) %>% inner_join(gene_tbl, by="probe_id") %>%
dplyr::select(-probe_id) %>% group_by(Entrez) %>% summarise_all(list(mean)) %>% as.data.frame()
rownames(gex_test) = gex_test$Entrez
gex_test = gex_test %>% dplyr::select(-Entrez)
## Conventional Gene Set Activity Analysis
# Gene set activity calculation
library(GSVAdata)## 필요한 패키지를 로딩중입니다: GSEABase## 필요한 패키지를 로딩중입니다: annotate## 필요한 패키지를 로딩중입니다: XML## 필요한 패키지를 로딩중입니다: graph##
## 다음의 패키지를 부착합니다: 'graph'## The following object is masked from 'package:XML':
##
## addNode## 필요한 패키지를 로딩중입니다: hgu95a.db##
library(ovltools)
data(c2BroadSets)
gs = lapply(c2BroadSets@.Data, function(x) x@geneIds)
gs_names = do.call(c, lapply(c2BroadSets@.Data, function(x) x@setName))
names(gs) = gs_names
es.gsva.train <- gsva(as.matrix(gex_train), gs, verbose=FALSE, parallel.sz=1, min.sz=3)
es.ss.train <- gsva(as.matrix(gex_train), gs, method="ssgsea", verbose=FALSE, parallel.sz=1, min.sz=3)
es.plage.train <- gsva(as.matrix(gex_train), gs, method="plage", verbose=FALSE, parallel.sz=1, min.sz=3)
es.gsva.test <- gsva(as.matrix(gex_test), gs, verbose=FALSE, parallel.sz=1, min.sz=3)
es.ss.test <- gsva(as.matrix(gex_test), gs, method="ssgsea", verbose=FALSE, parallel.sz=1, min.sz=3)
es.plage.test <- gsva(as.matrix(gex_test), gs, method="plage", verbose=FALSE, parallel.sz=1, min.sz=3)
# significance of Gene set activity
gsva_limma_res_train = limma_deg(es.gsva.train, label_train)
ss_limma_res_train = limma_deg(es.ss.train, label_train)
plage_limma_res_train = limma_deg(es.plage.train, label_train)
gsva_res_train = gsva_limma_res_train[gsva_limma_res_train$adj.P.Val < 0.05,]
ss_res_train = ss_limma_res_train[ss_limma_res_train$adj.P.Val < 0.05,]
plage_res_train = plage_limma_res_train[plage_limma_res_train$adj.P.Val < 0.05,]
gsva_train_gs = rownames(gsva_res_train)[gsva_res_train$P.Value < 0.05]
ss_train_gs = rownames(ss_res_train)[ss_res_train$P.Value < 0.05]
plage_train_gs = rownames(plage_res_train)[plage_res_train$P.Value < 0.05]
gsva_limma_res_test = limma_deg(es.gsva.test, label_test)
ss_limma_res_test = limma_deg(es.ss.test, label_test)
plage_limma_res_test = limma_deg(es.plage.test, label_test)
gsva_res_test = gsva_limma_res_test[gsva_limma_res_test$adj.P.Val < 0.05,]
ss_res_test = ss_limma_res_test[ss_limma_res_test$adj.P.Val < 0.05,]
plage_res_test = plage_limma_res_test[plage_limma_res_test$adj.P.Val < 0.05,]
gsva_test_gs = rownames(gsva_res_test)[gsva_res_test$P.Value < 0.05]
ss_test_gs = rownames(ss_res_test)[ss_res_test$P.Value < 0.05]
plage_test_gs = rownames(plage_res_test)[plage_res_test$P.Value < 0.05]
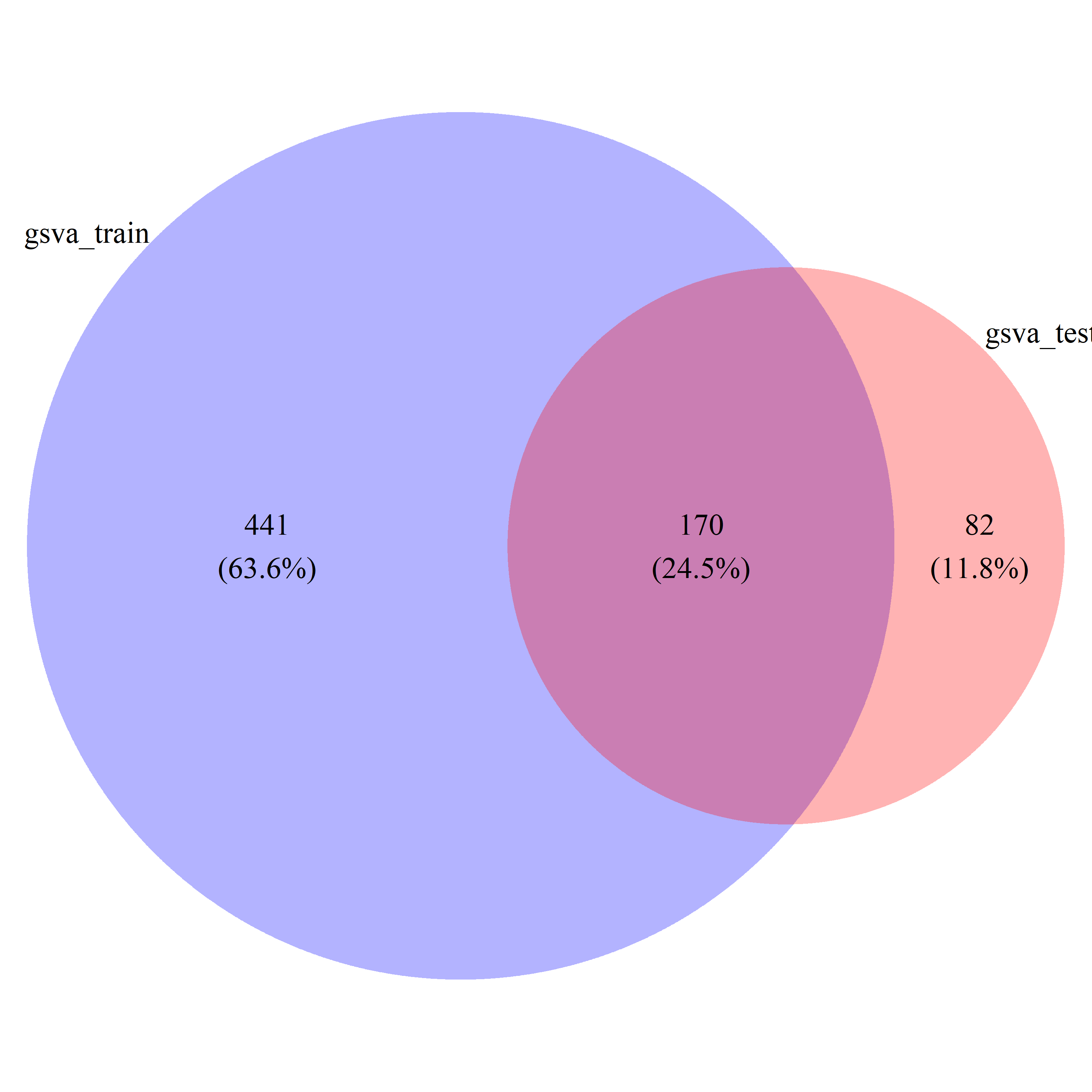
gsva_common_gs = intersect(gsva_train_gs, gsva_test_gs)
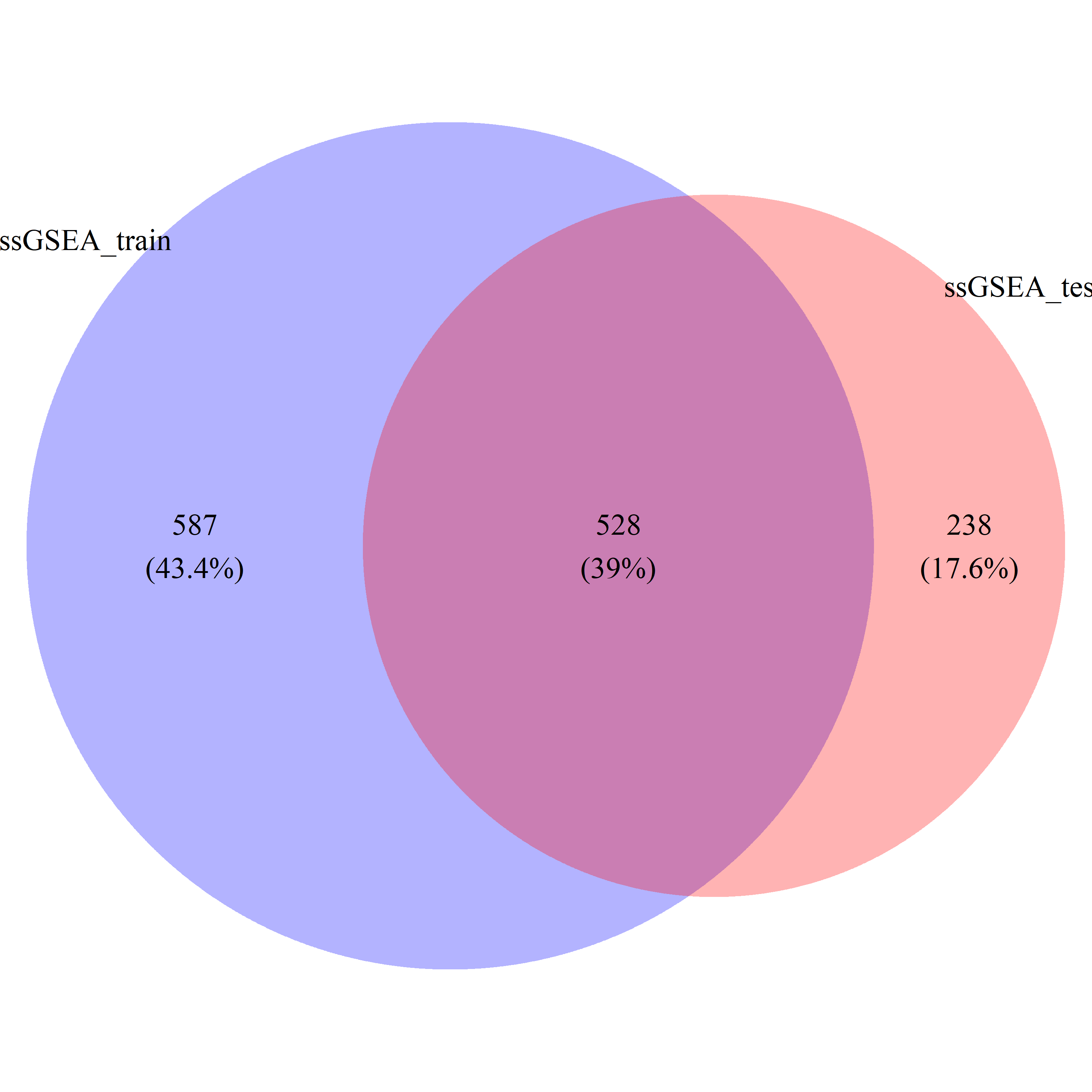
ss_common_gs = intersect(ss_train_gs, ss_test_gs)
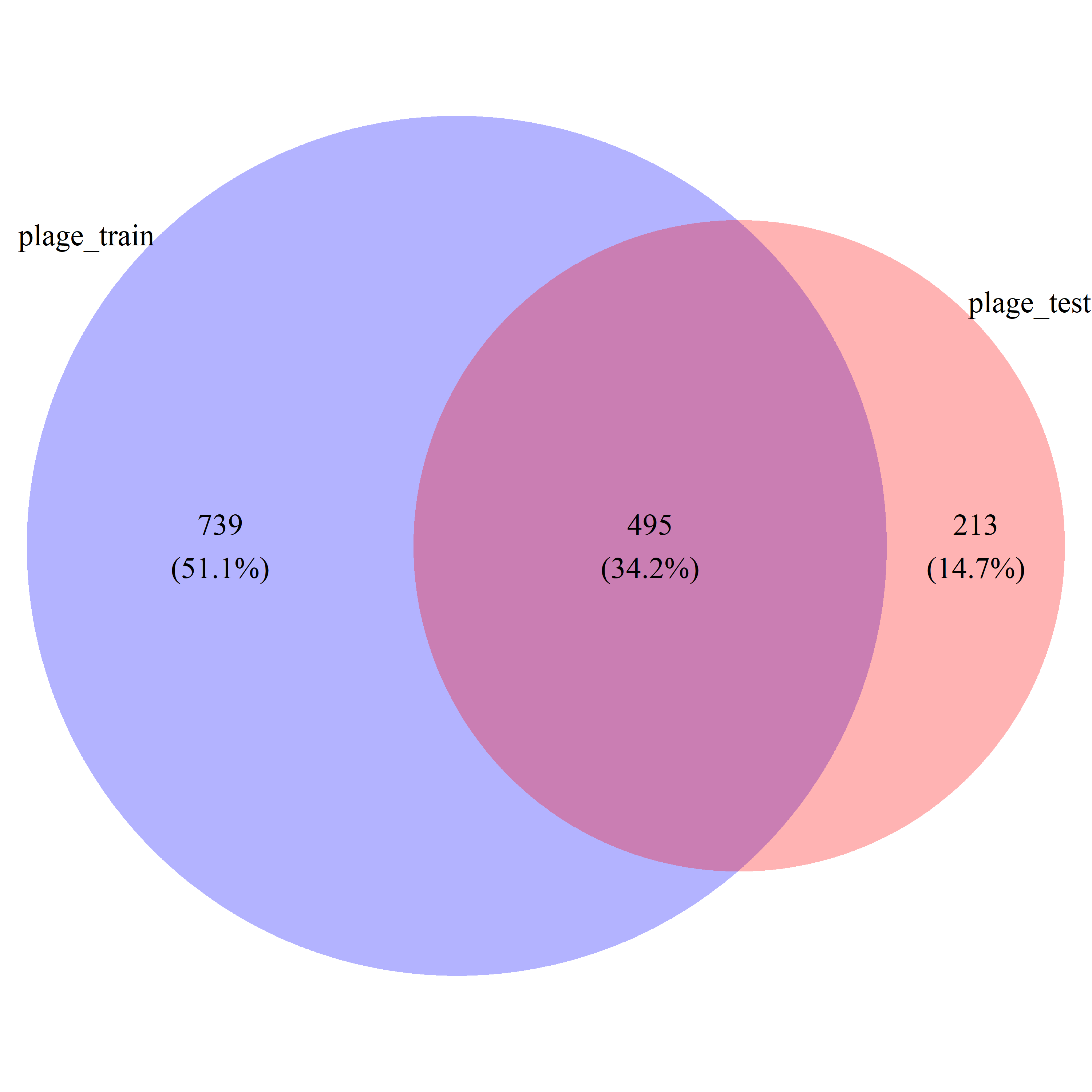
plage_common_gs = intersect(plage_train_gs, plage_test_gs)
gsva_all_gs = union(gsva_train_gs, gsva_test_gs)
ss_all_gs = union(ss_train_gs, ss_test_gs)
plage_all_gs = union(plage_train_gs, plage_test_gs)Differentially Active Gene Set Analysis with ovltools
## GSA based on ovl.test, Train case
# min.sz=3
# res = list()
# for(i in 1:length(gs)){
# if (i %% 30 == 0) message(paste(i,"/",length(gs)))
# tmp_dt = gex_train[rownames(gex_train) %in% gs[[i]],]
#
# if(dim(tmp_dt)[1] < min.sz) next
# tmp_dt = t(tmp_dt)
# pc = prcomp(tmp_dt, center=T, scale.=T)
# pc_vec = tmp_dt %*% pc$rotation[,1]
# tmp_res = ovltools::ovl.test(pc_vec[label_train==levels(label_train)[1]], pc_vec[label_train==levels(label_train)[2]], method = 'hist')
# res[[i]] = c(gs_name = names(gs)[[i]], tmp_res)
# }
# save(res, file="../data/ovl_dags_train_res_ovl_pca.RData")
load("../data/ovl_dags_train_res_ovl_pca.RData")
ovl_train_res = do.call(rbind, res)
ovl_train_res = data.frame(ovl_train_res)
ovl_train_res = ovl_train_res %>% arrange(pval)
ovl_train_gs = ovl_train_res$gs_name[ovl_train_res$pval<0.05]
## GSA based on ovl.test, Test case
# min.sz=3
# res = list()
# for(i in 1:length(gs)){
# if (i %% 30 == 0) message(paste(i,"/",length(gs)))
# tmp_dt = gex_test[rownames(gex_test) %in% gs[[i]],]
#
# if(dim(tmp_dt)[1] < min.sz) next
#
# tmp_dt = t(tmp_dt)
# pc = prcomp(tmp_dt, center=T, scale.=T)
# pc_vec = tmp_dt %*% pc$rotation[,1]
# tmp_res = ovltools::ovl.test(pc_vec[label_test==levels(label_test)[1]], pc_vec[label_test==levels(label_test)[2]], method = 'hist')
# res[[i]] = c(gs_name = names(gs)[[i]], tmp_res)
# }
# save(res, file="../data/ovl_dags_test_res_ovl_pca.RData")
load("../data/ovl_dags_test_res_ovl_pca.RData")
ovl_test_res = do.call(rbind, res)
ovl_test_res = data.frame(ovl_test_res)
ovl_test_res = ovl_test_res %>% arrange(pval)
ovl_test_gs = ovl_test_res$gs_name[ovl_test_res$pval<0.05]
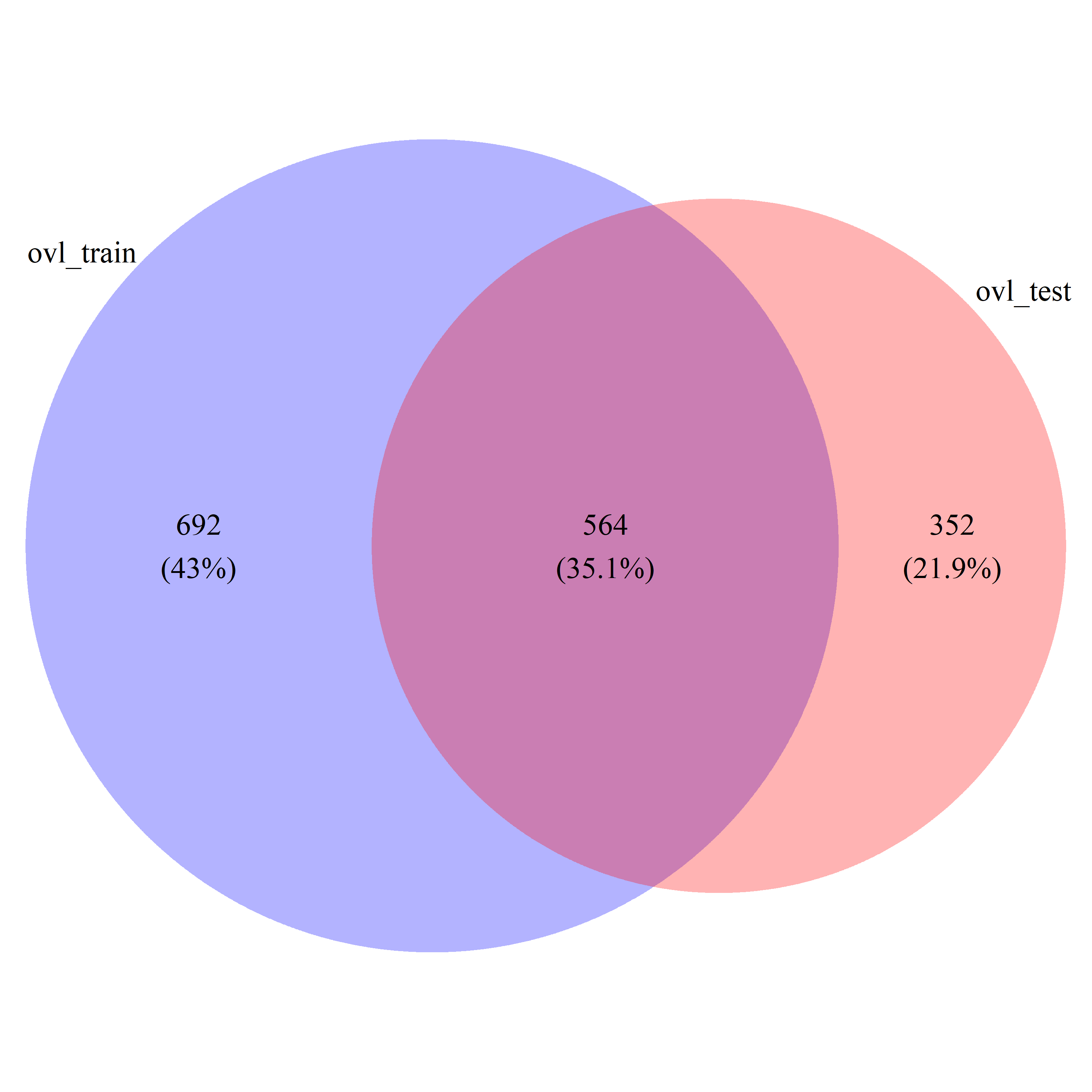
ovl_common_gs = intersect(ovl_train_gs, ovl_test_gs)
ovl_all_gs = union(ovl_train_gs, ovl_test_gs)Draw Venn diagram
# suppressPackageStartupMessages(library(VennDiagram))
# draw_venn(list(gsva_train_gs, gsva_test_gs), cat.name=c("gsva_train", "gsva_test"), f.name="../fig/golub_dags_gsva_VennDiagram.png")
# draw_venn(list(ss_train_gs, ss_test_gs), cat.name=c("ssGSEA_train", "ssGSEA_test"), f.name="../fig/golub_dags_ssgsea_VennDiagram.png")
# draw_venn(list(plage_train_gs, plage_test_gs), cat.name=c("plage_train", "plage_test"), f.name="../fig/golub_dags_plage_VennDiagram.png")
# draw_venn(list(ovl_train_gs, ovl_test_gs), cat.name=c("ovl_train", "ovl_test"), f.name="../fig/golub_dags_ovltools_VennDiagram.png")
#
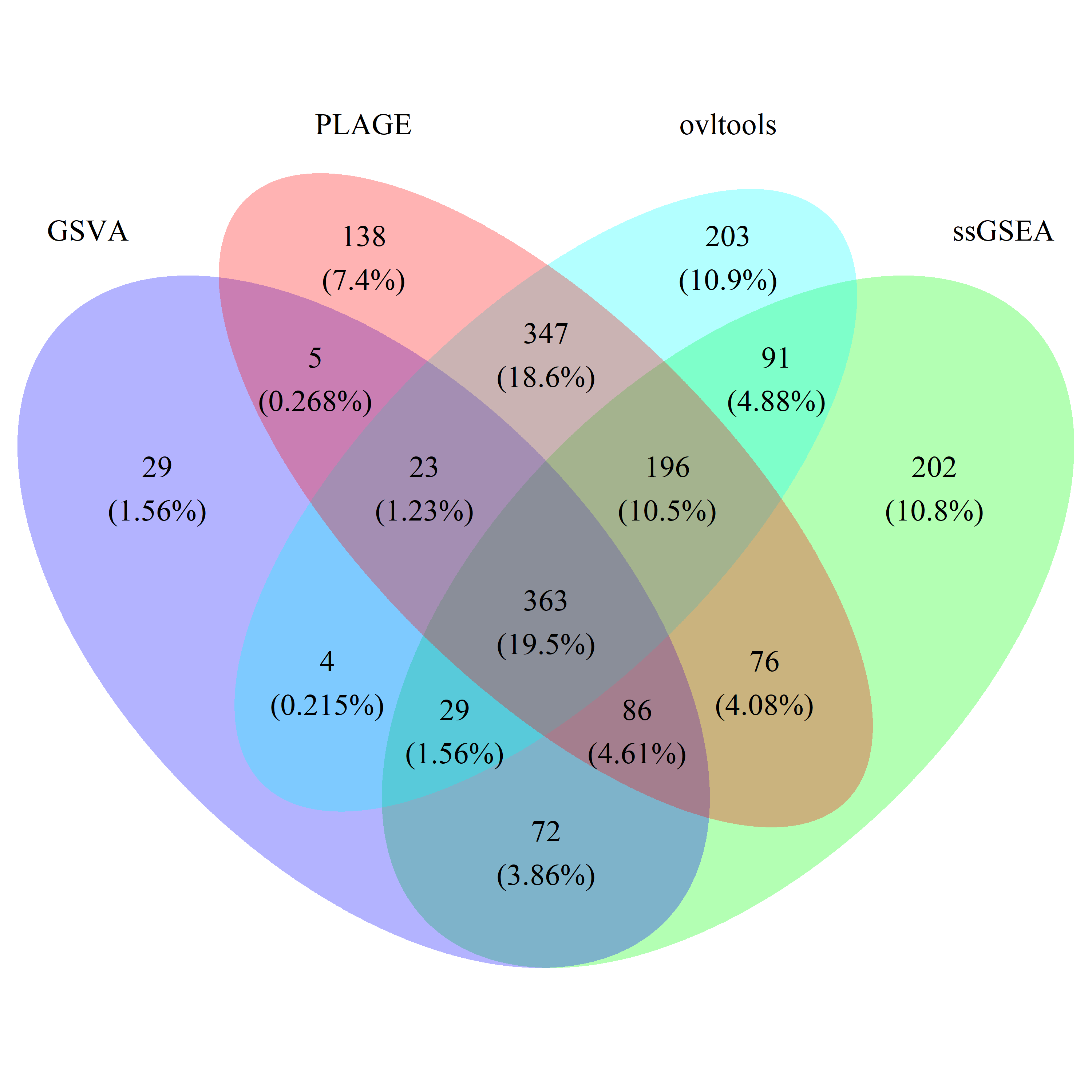
# draw_venn(list(gsva_train_gs, ss_train_gs, plage_train_gs, ovl_train_gs),
# cat.name=c("GSVA", "ssGSEA", "PLAGE", "ovltools"), f.name="../fig/golub_train_dags_VennDiagram.png")
#
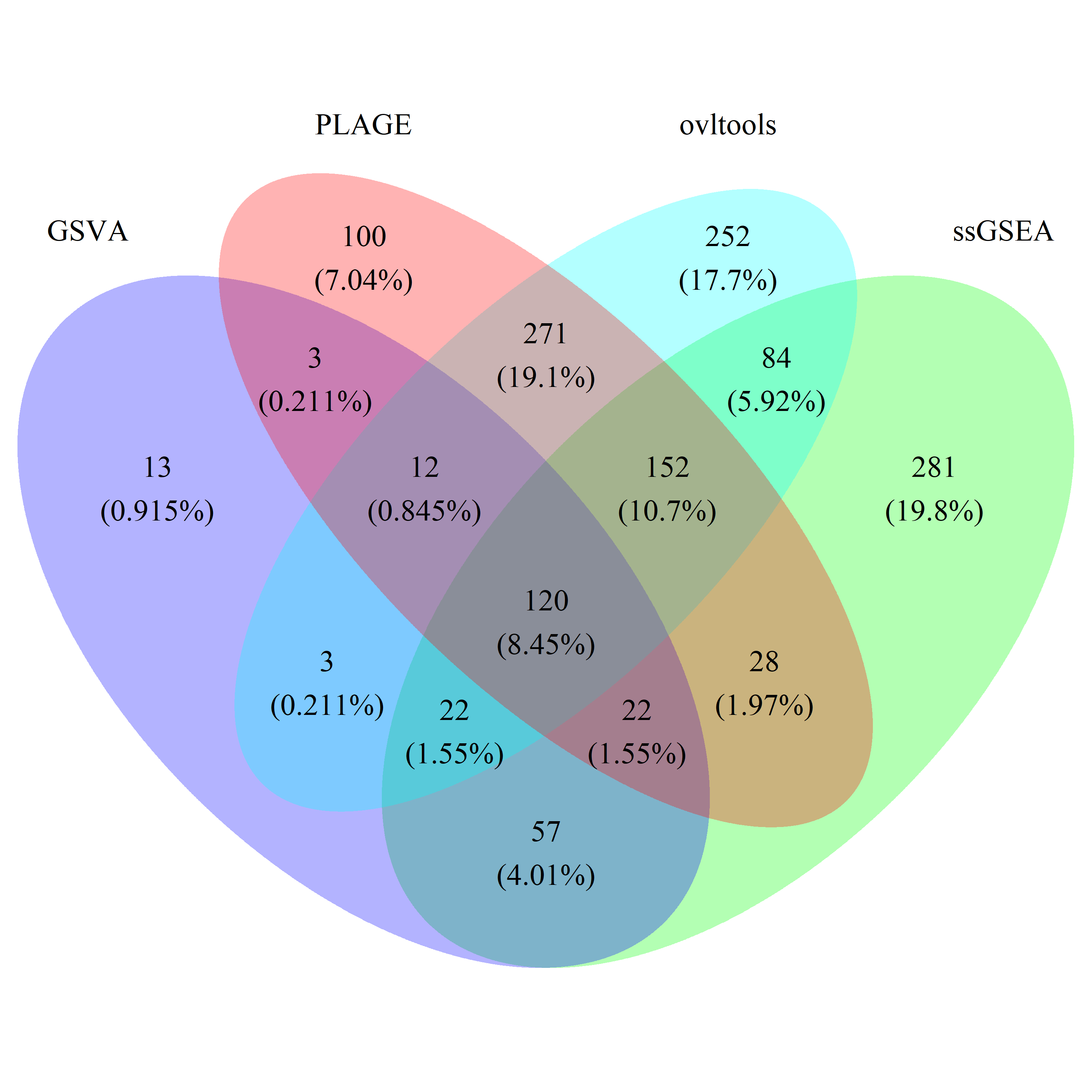
# draw_venn(list(gsva_test_gs, ss_test_gs, plage_test_gs, ovl_test_gs),
# cat.name=c("GSVA", "ssGSEA", "PLAGE", "ovltools"), f.name="../fig/golub_test_dags_VennDiagram.png")
#
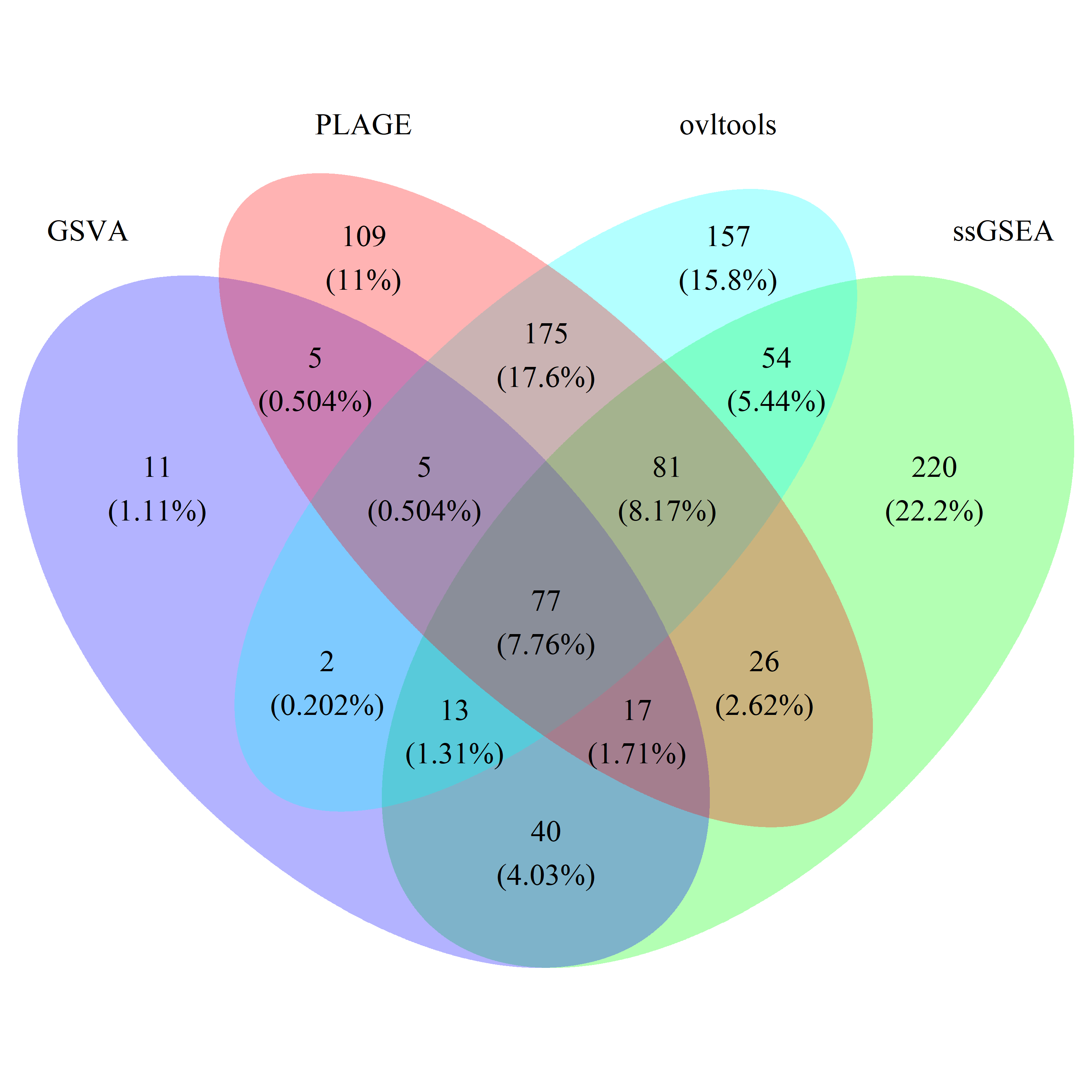
# draw_venn(list(gsva_common_gs, ss_common_gs, plage_common_gs, ovl_common_gs),
# cat.name=c("GSVA", "ssGSEA", "PLAGE", "ovltools"), f.name="../fig/golub_common_dags_VennDiagram.png")
#
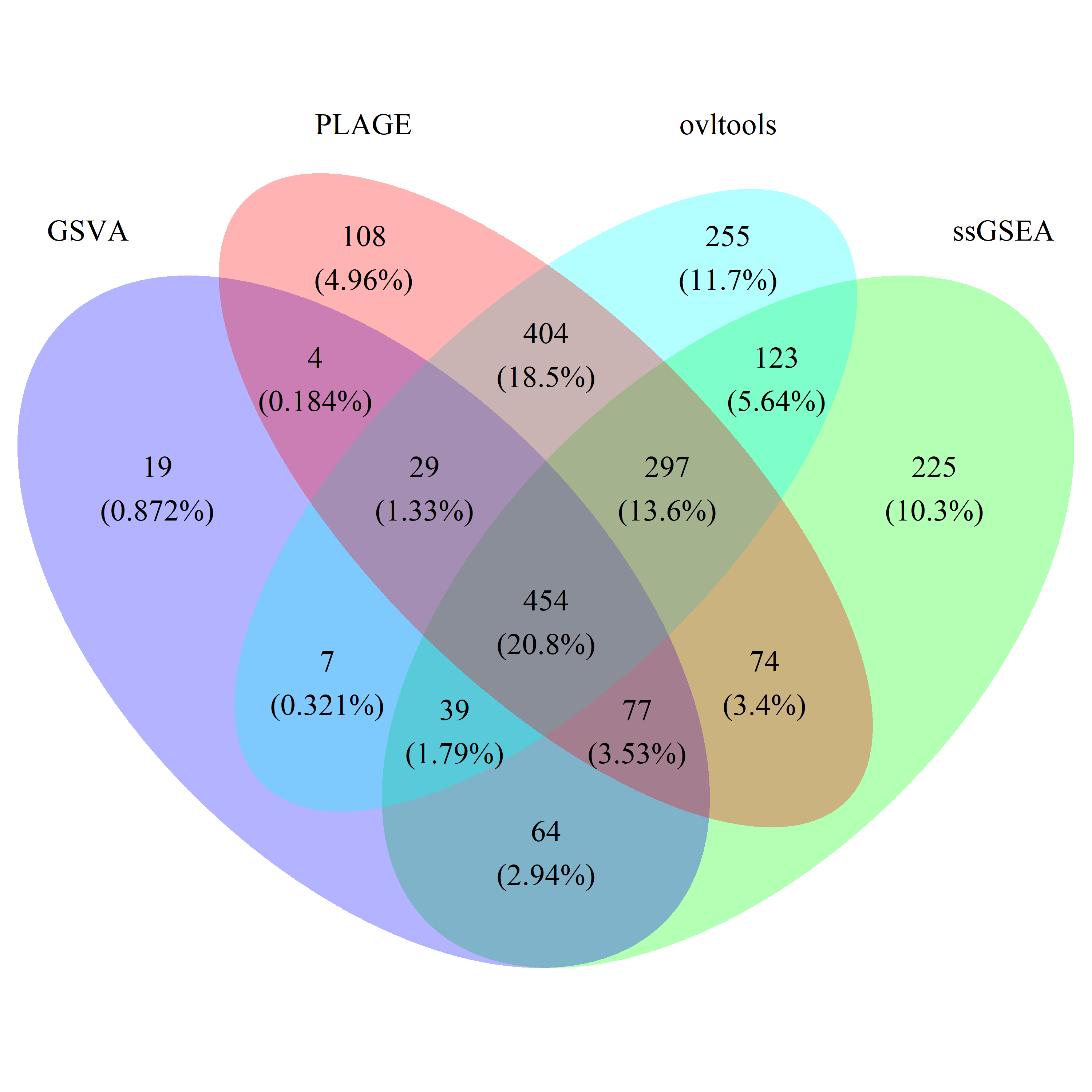
# draw_venn(list(gsva_all_gs, ss_all_gs, plage_all_gs, ovl_all_gs),
# cat.name=c("GSVA", "ssGSEA", "PLAGE", "ovltools"), f.name="../fig/golub_all_dags_VennDiagram.png")
# ovl_test_res %>% filter(gs_name %in% commonset) %>% arrange(desc(OVL))